⚪ My (failed) OpenCV antibiogram detection project
Trying to detect antibiotic disks and measure inhibition zones in OpenCV
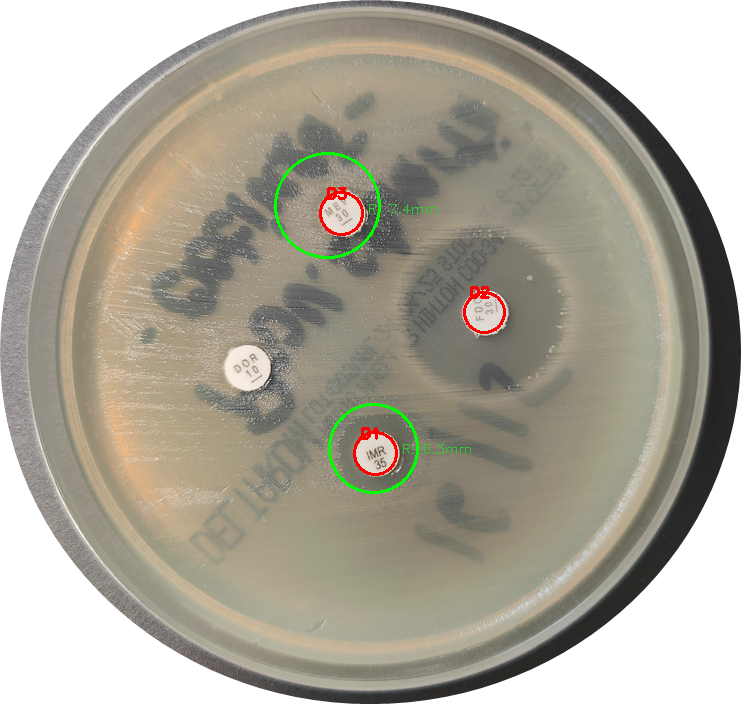
For a bit of a side project, I thought I’d try my hand at automating the reading of antibiograms using Python and OpenCV. The goal was to detect the antibiotic disks on an agar plate and then measure the diameter of the inhibition zones around them. Seemed like a classic computer vision task, and I was keen on trying it and learning more about the subject. They have a bunch of tutorials and good resources on their page. Definitely worth giving it a look. As far as I understand OpenCV is one of the oldest and coolest open-source computer vision projects, and a lot of people use it for a plethora of use cases.
Anyway, it turns out circles aren’t always as simple as they look, especially to a computer… 😑
👽 The plan
My approach involved a couple of main stages:
- Detecting the Antibiotic Disks: I figured SimpleBlobDetector in OpenCV would be a good starting point for finding the dark, circular antibiotic disks.
- Finding Inhibition Zones: Once a disk was found, the plan was to define a Region of Interest (ROI) around it, run Canny edge detection, and then use the Hough Circle Transform to find the clear circular zone of inhibition. Should work, right? Yeah, nah.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
import cv2
import numpy as np
# blob detection params
BLOB_MIN_CIRCULARITY = 0.7
BLOB_MIN_AREA = 400
BLOB_MAX_AREA = 16000
# Hough circle params
HC_DP = 1.2 # inverse ratio of accumulator resolution. 1 is full res.
HC_MIN_DIST_FACTOR = 2.0
HC_PARAM1 = 100
HC_PARAM2 = 18
HC_MIN_RADIUS_FACTOR = 1.1 # as factor of disk_radius_px
HC_MAX_RADIUS_FACTOR = 10.0
CANNY_THRESHOLD1 = 20
CANNY_THRESHOLD2 = 80
MAX_ALLOWED_OFFSET_FACTOR_HOUGH = 1.0
DISK_DIAMETER_MM = 6.0
DISK_RADIUS_MM = DISK_DIAMETER_MM / 2.0
image_path = "antibiogram_env/Poze/testE2cropped.png"
image = cv2.imread(image_path)
if image is None:
print(f"Error: Image not loaded. Check the path: {image_path}")
exit()
# converting to grayscale and apply gaussian blur
gray = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
blurred_plate = cv2.GaussianBlur(gray, (7, 7), 0)
blurred_roi_for_canny = cv2.GaussianBlur(gray, (5,5),0) # potentially for zone edge detection?
params = cv2.SimpleBlobDetector_Params()
params.filterByCircularity = True
params.minCircularity = BLOB_MIN_CIRCULARITY
params.filterByArea = True
params.minArea = BLOB_MIN_AREA
params.maxArea = BLOB_MAX_AREA
detector = cv2.SimpleBlobDetector_create(params)
keypoints = detector.detect(blurred_plate)
output_image = image.copy()
print("Detected disks and inhib zones:\n")
if not keypoints:
print("No disks detected")
else:
print(f"Found {len(keypoints)} disks.")
first_disk_canny_roi = None #
# loops through each detected disk (keypoint)
for i, kp in enumerate(keypoints):
x_disk, y_disk = int(kp.pt[0]), int(kp.pt[1])
disk_radius_px = int(kp.size / 2)
if disk_radius_px == 0:
print(f"Disk D{i+1} at ({x_disk},{y_disk}) has zero radius. Skipping.")
continue
# drawing the detected disks
cv2.circle(output_image, (x_disk, y_disk), disk_radius_px, (0, 0, 255), 2)
cv2.putText(output_image, f"D{i+1}", (x_disk - 15, y_disk - 15),
cv2.FONT_HERSHEY_SIMPLEX, 0.5, (0, 0, 255), 2)
# defines a ROI around the disk to search for the inhibition zone
roi_expansion_factor = HC_MAX_RADIUS_FACTOR + 1
roi_half_width = int(disk_radius_px * roi_expansion_factor)
x1_roi = max(x_disk - roi_half_width, 0)
x2_roi = min(x_disk + roi_half_width, gray.shape[1])
y1_roi = max(y_disk - roi_half_width, 0)
y2_roi = min(y_disk + roi_half_width, gray.shape[0])
roi_for_processing = blurred_roi_for_canny[y1_roi:y2_roi, x1_roi:x2_roi]
if roi_for_processing.size == 0 or roi_for_processing.shape[0] < 10 or roi_for_processing.shape[1] < 10:
print(f"Disk D{i+1}: ROI is too small or empty. Skipping.")
continue
# Canny Edge Detection within the ROI
edges_roi = cv2.Canny(roi_for_processing, CANNY_THRESHOLD1, CANNY_THRESHOLD2)
if i == 0: # this saves Canny output of the first disk's ROI for debugging
first_disk_canny_roi = edges_roi
# using Hough Circle transform to find the inhibition zone
min_dist_circles = int(disk_radius_px * HC_MIN_DIST_FACTOR)
hough_min_radius = int(disk_radius_px * HC_MIN_RADIUS_FACTOR)
hough_max_radius = int(disk_radius_px * HC_MAX_RADIUS_FACTOR)
if hough_min_radius >= hough_max_radius: # basic sanity check :))
hough_max_radius = hough_min_radius + disk_radius_px
print(f"\n--- Proc Disk D{i+1} at ({x_disk},{y_disk}), DiskRadiusPx: {disk_radius_px} ---")
circles = cv2.HoughCircles(edges_roi, cv2.HOUGH_GRADIENT, dp=HC_DP, minDist=min_dist_circles,
param1=HC_PARAM1, param2=HC_PARAM2,
minRadius=hough_min_radius, maxRadius=hough_max_radius)
best_hough_circle = None
if circles is not None:
circles = np.uint16(np.around(circles))
print(f" Found {circles.shape[1]} raw circle(s) for Disk D{i+1}.")
candidate_zones = []
for c_idx, c in enumerate(circles[0, :]):
cx_roi, cy_roi, radius_hough_px = c[0], c[1], c[2]
print(f" Raw C{c_idx}: CenterROI=({cx_roi},{cy_roi}), RadiusPx={radius_hough_px}")
# this checks concentricity with the original disk
disk_center_roi_x = x_disk - x1_roi
disk_center_roi_y = y_disk - y1_roi
max_allowed_offset_px = disk_radius_px * MAX_ALLOWED_OFFSET_FACTOR_HOUGH
dist_to_disk_center = np.sqrt((cx_roi - disk_center_roi_x)**2 + (cy_roi - disk_center_roi_y)**2)
if dist_to_disk_center > max_allowed_offset_px:
print(f" REJECT (Offset): Dist={dist_to_disk_center:.1f} > MaxOffset={max_allowed_offset_px:.1f}")
continue
# find radius in mm
inhib_zone_radius_mm = (radius_hough_px / disk_radius_px) * DISK_RADIUS_MM if disk_radius_px > 0 else 0
print(f" ACCEPT: RadiusMM={inhib_zone_radius_mm:.1f}, Offset={dist_to_disk_center:.1f}")
candidate_zones.append({
"radius_mm": inhib_zone_radius_mm,
"radius_px": radius_hough_px,
"center_roi": (cx_roi, cy_roi),
"offset": dist_to_disk_center
})
if candidate_zones:
# selects the "best" candidate (by looking at smallest offset)
candidate_zones.sort(key=lambda z: z["offset"])
best_hough_circle = candidate_zones[0]
if best_hough_circle:
# Converting ROI coordinates back to original img coords
abs_zone_center_x = best_hough_circle["center_roi"][0] + x1_roi
abs_zone_center_y = best_hough_circle["center_roi"][1] + y1_roi
# this draws the inhib zone
cv2.circle(output_image, (abs_zone_center_x, abs_zone_center_y),
best_hough_circle["radius_px"], (0, 255, 0), 2)
cv2.putText(output_image, f"R: {best_hough_circle['radius_mm']:.1f}mm",
(x_disk + disk_radius_px + 5, y_disk),
cv2.FONT_HERSHEY_SIMPLEX, 0.45, (50, 200, 50), 1)
print(f" Disk D{i+1} (at {x_disk},{y_disk}): InhibZ RADIUS = {best_hough_circle['radius_mm']:.1f} mm (Offset: {best_hough_circle['offset']:.1f}px)")
else:
print(f" Disk D{i+1} (at {x_disk},{y_disk}): No valid inhib zone circle found via Hough transform.")
# displaying the Canny edges for the first disk's ROI to debug better
if first_disk_canny_roi is not None:
cv2.imshow("Sample Canny Edge ROI (Disk D1)", first_disk_canny_roi)
else:
print("Could not generate a sample Canny ROI.")
# displaying the final image with detected disks and zones
cv2.imshow("Disks and Inhibition Zones", output_image)
cv2.waitKey(0)
cv2.destroyAllWindows()
I tried to find the antibiotic disks first using SimpleBlobDetector. Then, for each disk, the script defines a region of interest, applies Canny edge detection, and then uses HoughCircles to find the inhibition zone. There’s also logic to filter Hough circles based on concentricity with the disk.
Despite a lot of parameter tweaking (you can see all the BLOB_ and HC_ constants I was playing with), getting consistent and reliable detection of both the disks and, more importantly, the often faint edges of the inhibition zones, proved really tough. You can see how even in this pretty simple example the algorithm struggled to detect one of the disks as well as one of the larger inhibition zones, whilst falsely detecting a zone where there wasn’t any.
🤖 I’LL BE BACK
While traditional OpenCV is amazing, this kind of complex pattern recognition in variable biological images is often where Machine Learning and AI really shine these days.
My plan is to eventually come back to this antibiogram challenge, but next time I’ll be looking into AI-driven solutions. Maybe a dedicated object detection model (like YOLOv2 - no, not “you only live once”, calm down there millennials) trained on disks and zones, or perhaps leveraging some of the newer multimodal LLMs that are getting good at image interpretation.
When I do (and hopefully have a different story to tell), I’ll be sure to post an update. For now, this particular OpenCV quest is paused.